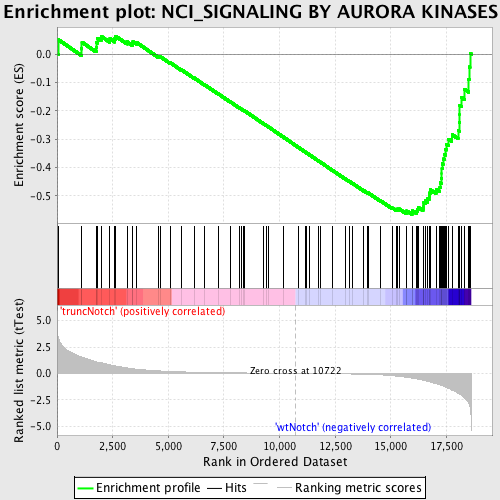
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_wtNotch.phenotype_truncNotch_versus_wtNotch.cls #truncNotch_versus_wtNotch |
| Phenotype | phenotype_truncNotch_versus_wtNotch.cls#truncNotch_versus_wtNotch |
| Upregulated in class | wtNotch |
| GeneSet | NCI_SIGNALING BY AURORA KINASES |
| Enrichment Score (ES) | -0.5659319 |
| Normalized Enrichment Score (NES) | -1.6264834 |
| Nominal p-value | 0.002793296 |
| FDR q-value | 0.2274387 |
| FWER p-Value | 0.884 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ARRB2 | 2060441 | 48 | 3.434 | 0.0515 | No | ||
| 2 | CDC25B | 6940102 | 1097 | 1.538 | 0.0191 | No | ||
| 3 | RELB | 1400048 | 1117 | 1.525 | 0.0421 | No | ||
| 4 | NFKBIA | 1570152 | 1757 | 1.101 | 0.0250 | No | ||
| 5 | LCK | 3360142 | 1776 | 1.087 | 0.0411 | No | ||
| 6 | NFKB2 | 2320670 | 1817 | 1.069 | 0.0558 | No | ||
| 7 | RHOA | 580142 5900131 5340450 | 1979 | 0.998 | 0.0628 | No | ||
| 8 | NFKB1 | 5420358 | 2374 | 0.822 | 0.0545 | No | ||
| 9 | OAZ1 | 110181 | 2594 | 0.724 | 0.0541 | No | ||
| 10 | PRKACA | 2640731 4050048 | 2625 | 0.708 | 0.0636 | No | ||
| 11 | GADD45A | 2900717 | 3144 | 0.512 | 0.0437 | No | ||
| 12 | TP53 | 6130707 | 3380 | 0.442 | 0.0380 | No | ||
| 13 | RACGAP1 | 3990162 6620736 | 3408 | 0.437 | 0.0434 | No | ||
| 14 | TNFAIP3 | 2900142 | 3560 | 0.405 | 0.0417 | No | ||
| 15 | VIM | 20431 | 4577 | 0.235 | -0.0095 | No | ||
| 16 | PRKCA | 6400551 | 4645 | 0.229 | -0.0095 | No | ||
| 17 | KIF20A | 2650050 | 5090 | 0.183 | -0.0305 | No | ||
| 18 | MYLK | 4010600 7000364 | 5581 | 0.144 | -0.0547 | No | ||
| 19 | ERC1 | 1580170 2630717 6110762 6840280 | 6154 | 0.111 | -0.0838 | No | ||
| 20 | BRCA1 | 4780669 | 6644 | 0.090 | -0.1088 | No | ||
| 21 | AURKC | 4060687 | 7232 | 0.069 | -0.1394 | No | ||
| 22 | ARHGEF7 | 1690441 | 7815 | 0.054 | -0.1700 | No | ||
| 23 | PSMA3 | 5900047 7040161 | 8176 | 0.045 | -0.1887 | No | ||
| 24 | SYK | 6940133 | 8296 | 0.042 | -0.1944 | No | ||
| 25 | DES | 1450341 | 8386 | 0.041 | -0.1986 | No | ||
| 26 | SGOL1 | 1980075 5220092 6020711 | 8402 | 0.040 | -0.1988 | No | ||
| 27 | CBX5 | 3830072 6290167 | 9256 | 0.024 | -0.2444 | No | ||
| 28 | PIK3R1 | 4730671 | 9411 | 0.022 | -0.2524 | No | ||
| 29 | NPM1 | 4730427 | 9492 | 0.020 | -0.2564 | No | ||
| 30 | BUB1 | 5390270 | 10159 | 0.009 | -0.2922 | No | ||
| 31 | XPO1 | 540707 | 10863 | -0.002 | -0.3301 | No | ||
| 32 | FBXW11 | 6450632 | 11158 | -0.007 | -0.3458 | No | ||
| 33 | PAK1 | 4540315 | 11187 | -0.008 | -0.3472 | No | ||
| 34 | AKT1 | 5290746 | 11366 | -0.010 | -0.3566 | No | ||
| 35 | KIF2C | 6940082 | 11750 | -0.018 | -0.3770 | No | ||
| 36 | RAN | 2260446 4590647 | 11827 | -0.019 | -0.3808 | No | ||
| 37 | SRC | 580132 | 12374 | -0.031 | -0.4098 | No | ||
| 38 | JUB | 5860711 5900500 | 12953 | -0.047 | -0.4403 | No | ||
| 39 | TDRD7 | 6550520 | 13121 | -0.052 | -0.4484 | No | ||
| 40 | KLHL13 | 6590731 | 13297 | -0.059 | -0.4570 | No | ||
| 41 | IKBKG | 3450092 3840377 6590592 | 13790 | -0.082 | -0.4822 | No | ||
| 42 | ATM | 3610110 4050524 | 13930 | -0.090 | -0.4883 | No | ||
| 43 | PIK3CA | 6220129 | 13980 | -0.093 | -0.4895 | No | ||
| 44 | RELA | 3830075 | 14534 | -0.138 | -0.5172 | No | ||
| 45 | TNFRSF1A | 1090390 6520735 | 15074 | -0.215 | -0.5429 | No | ||
| 46 | PPP1CC | 6380300 2510647 | 15240 | -0.251 | -0.5478 | No | ||
| 47 | NOD2 | 2510050 | 15292 | -0.261 | -0.5465 | No | ||
| 48 | REL | 360707 | 15372 | -0.279 | -0.5463 | No | ||
| 49 | EVI5 | 6380040 | 15696 | -0.367 | -0.5580 | No | ||
| 50 | MDM2 | 3450053 5080138 | 15723 | -0.375 | -0.5535 | No | ||
| 51 | BCL10 | 2360397 | 15955 | -0.455 | -0.5588 | Yes | ||
| 52 | AURKB | 5890739 | 15969 | -0.458 | -0.5523 | Yes | ||
| 53 | SMC4 | 5910240 | 16168 | -0.524 | -0.5547 | Yes | ||
| 54 | CPEB1 | 3190524 | 16189 | -0.533 | -0.5474 | Yes | ||
| 55 | SMC2 | 4810133 | 16227 | -0.548 | -0.5408 | Yes | ||
| 56 | STMN1 | 1990717 | 16449 | -0.636 | -0.5427 | Yes | ||
| 57 | TNF | 6650603 | 16480 | -0.652 | -0.5340 | Yes | ||
| 58 | TRAF6 | 4810292 6200132 | 16483 | -0.654 | -0.5238 | Yes | ||
| 59 | MALT1 | 4670292 | 16550 | -0.685 | -0.5166 | Yes | ||
| 60 | BIRC2 | 2940435 4730020 | 16663 | -0.757 | -0.5107 | Yes | ||
| 61 | BIRC5 | 110408 580014 1770632 | 16722 | -0.790 | -0.5014 | Yes | ||
| 62 | CENPA | 5080154 | 16744 | -0.803 | -0.4899 | Yes | ||
| 63 | TACC3 | 5130592 | 16768 | -0.814 | -0.4784 | Yes | ||
| 64 | PPP2R5D | 4010156 380408 5550112 5670162 | 17034 | -0.982 | -0.4772 | Yes | ||
| 65 | BCL3 | 3990440 | 17204 | -1.061 | -0.4696 | Yes | ||
| 66 | INCENP | 520593 | 17229 | -1.082 | -0.4539 | Yes | ||
| 67 | GSK3B | 5360348 | 17269 | -1.119 | -0.4384 | Yes | ||
| 68 | MAP3K14 | 5890435 | 17297 | -1.137 | -0.4219 | Yes | ||
| 69 | AURKA | 780537 | 17299 | -1.140 | -0.4040 | Yes | ||
| 70 | MAPK14 | 5290731 | 17315 | -1.150 | -0.3867 | Yes | ||
| 71 | RIPK2 | 5050072 6290632 | 17351 | -1.187 | -0.3699 | Yes | ||
| 72 | IKBKB | 6840072 | 17404 | -1.233 | -0.3533 | Yes | ||
| 73 | CYLD | 6590079 | 17461 | -1.276 | -0.3362 | Yes | ||
| 74 | CDCA8 | 2340286 6980019 | 17502 | -1.320 | -0.3176 | Yes | ||
| 75 | KIF23 | 5570112 | 17597 | -1.409 | -0.3005 | Yes | ||
| 76 | NDC80 | 4120465 | 17749 | -1.583 | -0.2837 | Yes | ||
| 77 | NCL | 2360463 4540279 | 18025 | -1.866 | -0.2692 | Yes | ||
| 78 | UBE2D3 | 3190452 | 18074 | -1.915 | -0.2416 | Yes | ||
| 79 | RASA1 | 1240315 | 18093 | -1.945 | -0.2120 | Yes | ||
| 80 | NSUN2 | 2120193 | 18099 | -1.961 | -0.1814 | Yes | ||
| 81 | CUL3 | 1850520 | 18170 | -2.066 | -0.1526 | Yes | ||
| 82 | TPX2 | 6420324 | 18310 | -2.306 | -0.1238 | Yes | ||
| 83 | CHUK | 7050736 | 18503 | -2.821 | -0.0898 | Yes | ||
| 84 | NDEL1 | 2370465 | 18522 | -2.900 | -0.0451 | Yes | ||
| 85 | NCAPH | 6220435 | 18560 | -3.183 | 0.0030 | Yes |

